 |
GROMACSMain Table of Contents |
VERSION 4.0
Sun 18 Jan 2009
| VERSION 4.0 |
This is a flow chart of a typical GROMACS MD run of a protein in a box of water. A more detailed example is available in the Getting Started section. Several steps of energy minimization may be necessary, these consist of cycles: grompp -> mdrun.
| eiwit.pdb |  |
|||||||
| Generate a GROMACS topology | pdb2gmx |  |
||||||
 |
 |
|||||||
| conf.gro | topol.top | |||||||
 |
     |
|||||||
| Enlarge the box | editconf |  |
||||||
 |
||||||||
| conf.gro | ||||||||
 |
||||||||
| Solvate protein | genbox |  |
||||||
 |
 |
|||||||
| conf.gro | topol.top | |||||||
| grompp.mdp |  |
 |
 |
|||||
| Generate mdrun input file | grompp |  |
||||||
 |
 |
Continuation | ||||||
| topol.tpr | state.cpt | |||||||
 |
  |
|||||||
| Run the simulation (EM or MD) | mdrun | |||||||
 |
 |
|||||||
| traj.xtc / traj.trr | ener.edr | |||||||
 |
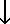 |
|||||||
| Analysis | g_... ngmx |
g_energy |  |
|||||