|
Gromacs
2025-dev-20240725-9868da9
|
|
Gromacs
2025-dev-20240725-9868da9
|
 Collaboration diagram for Framework for Trajectory Analysis (trajectoryanalysis):
Collaboration diagram for Framework for Trajectory Analysis (trajectoryanalysis):Provides functionality for implementing trajectory analysis modules.
This module implements a framework for implementing flexible trajectory analysis routines. It provides a base class for implementing analysis as reusable modules that can be used from different contexts and can also support per-frame parallelization. It integrally uses functionality from the following modules:
The main interface of this module is the gmx::TrajectoryAnalysisModule class. Analysis modules should derive from this class, and override the necessary virtual methods to provide the actual initialization and analysis routines. Classes gmx::TrajectoryAnalysisSettings and gmx::TopologyInformation (in addition to classes declared in the above-mentioned modules) are used to pass information to and from these methods. gmx::TrajectoryAnalysisModuleData can be used in advanced scenarios where the tool requires custom thread-local data for parallel analysis.
The sequence charts below provides an overview of how the trajectory analysis modules typically interact with other components. The first chart provides an overview of the call sequence of the most important methods in gmx::TrajectoryAnalysisModule. There is a runner, which is responsible for doing the work that is shared between all trajectory analysis (such as reading the trajectory and processing selections). The runner then calls different methods in the analysis module at appropriate points to perform the module-specific tasks. The analysis module is responsible for creating and managing gmx::AnalysisData objects, and the chart shows the most important interactions with this module as well. However, the runner takes responsibility of calling gmx::AnalysisData::finishFrameSerial(). Interactions with options (for command-line option processing) and selections is not shown for brevity: see Extensible Handling of Options (options) for an overview of how options work, and the second chart for a more detailed view of how selections are accessed from an analysis module.
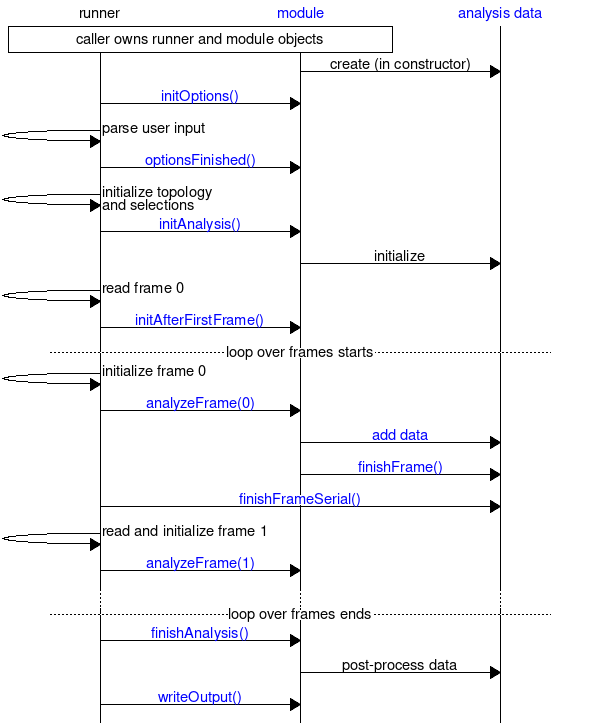
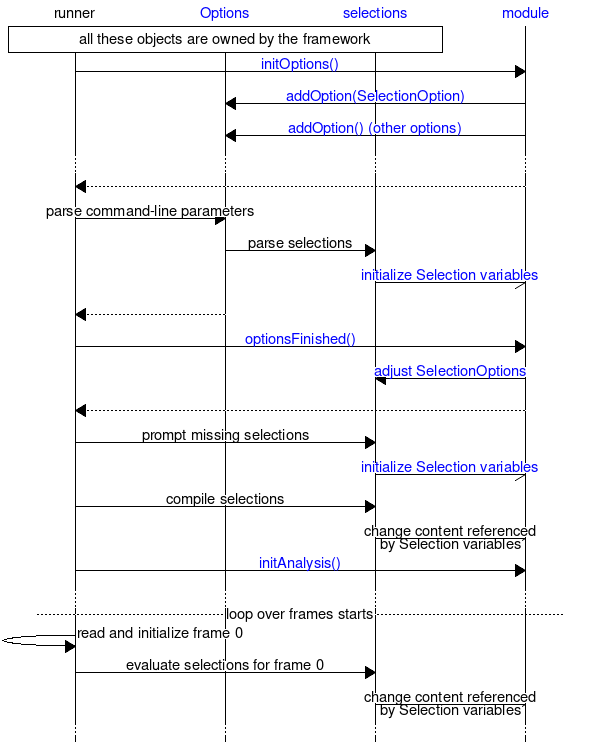
The second chart below shows the interaction with selections and options with focus on selection options. The gmx::TrajectoryAnalysisModule object creates one or more gmx::Selection variables, and uses gmx::SelectionOption to indicate them as the destination for selections. This happens in gmx::TrajectoryAnalysisModule::initOptions(). After the options have been parsed (includes parsing any options present on the command-line or read from files, but not those provided interactively), gmx::TrajectoryAnalysisModule::optionsFinished() can adjust the selections using gmx::SelectionOptionInfo. This is done like this to allow the analysis module to influence the interactive prompt of selections based on what command-line options were given. After optionsFinished() returns, the interactive selection prompt is presented if necessary. After this point, all access to selections from the analysis module is through the gmx::Selection variables: the runner is responsible for calling methods in the selection library, and these methods update the content referenced by the gmx::Selection variables. See documentation of gmx::TrajectoryAnalysisModule for details of what the selections contain at each point.
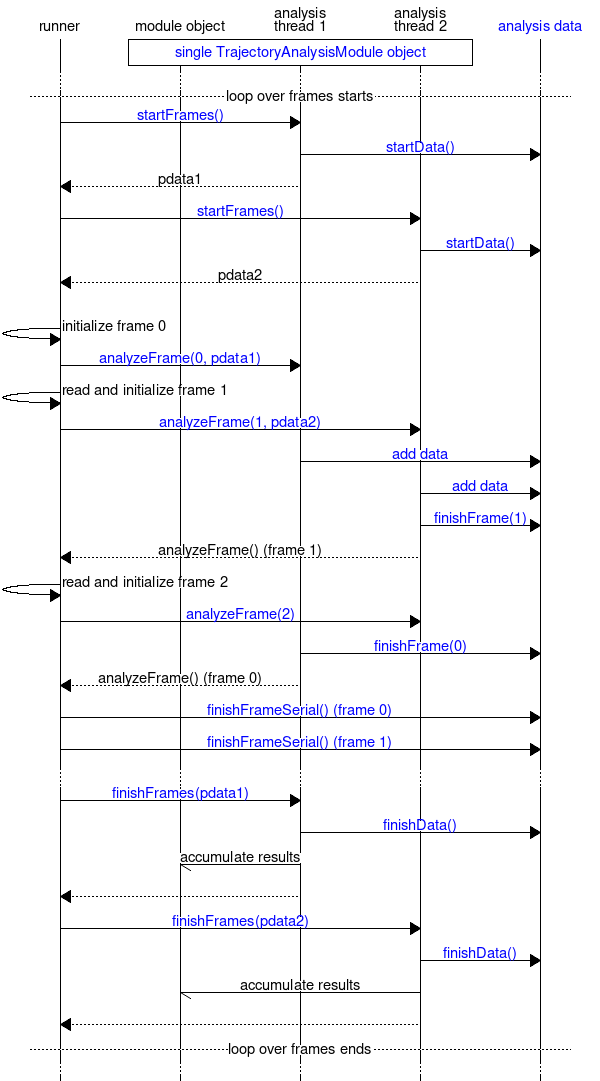
The final chart shows the flow within the frame loop in the case of parallel (threaded) execution and the interaction with the Parallelizable Handling of Output Data (analysisdata) module in this case. Although parallelization has not yet been implemented, it has influenced the design and needs to be understood if one wants to write modules that can take advantage of the parallelization once it gets implemented. The parallelization takes part over frames: analyzing a single frame is one unit of work. When the frame loop is started, gmx::TrajectoryAnalysisModule::startFrames() is called for each thread, and initializes an object that contains thread-local data needed during the analysis. This includes selection information, gmx::AnalysisDataHandle objects, and possibly other module-specific variables. Then, the runner reads the frames in sequence and passes the work into the different threads, together with the appropriate thread-local data object. The gmx::TrajectoryAnalysisModule::analyzeFrame() calls are only allowed to modify the thread-local data object; everything else is read-only. For any output, they pass the information to gmx::AnalysisData, which together with the runner takes care of ordering the data from different frames such that it gets processed in the right order. When all frames are analyzed, gmx::TrajectoryAnalysisModule::finishFrames() is called for each thread-local data object to destroy them and to accumulate possible results from them into the main gmx::TrajectoryAnalysisModule object. Note that in the diagram, some part of the work attributed for the runner (e.g., evaluating selections) will actually be carried out in the analysis threads before gmx::TrajectoryAnalysisModule::analyzeFrame() gets called.
In addition to the framework for defining analysis modules, this module also provides gmx::TrajectoryAnalysisCommandLineRunner, which implements a command-line program that runs a certain analysis module.
Internally, the module also defines a set of trajectory analysis modules that can currently be accessed only through gmx::registerTrajectoryAnalysisModules.
For an example of how to implement an analysis tool using the framework, see template.cpp.
Classes | |
| class | gmx::TrajectoryAnalysisModule::Impl |
| Private implementation class for TrajectoryAnalysisModule. More... | |
| class | gmx::anonymous_namespace{analysismodule.cpp}::TrajectoryAnalysisModuleDataBasic |
| Basic thread-local trajectory analysis data storage class. More... | |
| class | gmx::analysismodules::anonymous_namespace{angle.cpp}::AnglePositionIterator |
| Helper to encapsulate logic for looping over input selections. More... | |
| class | gmx::analysismodules::anonymous_namespace{freevolume.cpp}::FreeVolume |
| Class used to compute free volume in a simulations box. More... | |
| class | gmx::analysismodules::anonymous_namespace{pairdist.cpp}::PairDistance |
Implements gmx pairdist trajectory analysis module. More... | |
| class | gmx::analysismodules::anonymous_namespace{pairdist.cpp}::PairDistanceModuleData |
| Temporary memory for use within a single-frame calculation. More... | |
| class | gmx::analysismodules::anonymous_namespace{rdf.cpp}::Rdf |
Implements gmx rdf trajectory analysis module. More... | |
| class | gmx::analysismodules::anonymous_namespace{rdf.cpp}::RdfModuleData |
| Temporary memory for use within a single-frame calculation. More... | |
| struct | gmx::analysismodules::anonymous_namespace{sasa.cpp}::t_conect |
| Tracks information on two nearest neighbors of a single surface dot. More... | |
| class | gmx::analysismodules::anonymous_namespace{sasa.cpp}::Sasa |
Implements gmx sas trajectory analysis module. More... | |
| class | gmx::analysismodules::anonymous_namespace{sasa.cpp}::SasaModuleData |
| Temporary memory for use within a single-frame calculation. More... | |
| class | gmx::SurfaceAreaCalculator |
| Computes surface areas for a group of atoms/spheres. More... | |
| class | gmx::UnionFinder |
| Union-find data structure for keeping track of disjoint sets. More... | |
| class | gmx::MappedUnionFinder |
| Extension of UnionFind that supports non-consecutive integer indices as items. More... | |
| class | gmx::TrajectoryAnalysisRunnerCommon |
| Implements common trajectory analysis runner functionality. More... | |
| class | gmx::test::AbstractTrajectoryAnalysisModuleTestFixture |
| Test fixture for trajectory analysis modules. More... | |
| class | gmx::test::TrajectoryAnalysisModuleTestFixture< ModuleInfo > |
| Test fixture for a trajectory analysis module. More... | |
| class | gmx::TrajectoryAnalysisModuleData |
| Base class for thread-local data storage during trajectory analysis. More... | |
| class | gmx::TrajectoryAnalysisModule |
| Base class for trajectory analysis modules. More... | |
| class | gmx::TrajectoryAnalysisSettings |
| Trajectory analysis module configuration object. More... | |
| class | gmx::TrajectoryAnalysisCommandLineRunner |
| Runner for command-line trajectory analysis tools. More... | |
| class | gmx::TopologyInformation |
| Topology information available to a trajectory analysis module. More... | |
Enumerations | |
| enum | gmx::analysismodules::anonymous_namespace{pairdist.cpp}::DistanceType : int { Min, Max, Count } |
Enum value to store the selected value for -type. | |
| enum | gmx::analysismodules::anonymous_namespace{pairdist.cpp}::GroupType : int { All, Residue, Molecule, None, Count } |
Enum value to store the selected value for -refgrouping/-selgrouping. | |
| enum | gmx::analysismodules::anonymous_namespace{rdf.cpp}::Normalization : int { Rdf, NumberDensity, None, Count } |
| Normalization for the computed distribution. | |
| enum | gmx::analysismodules::anonymous_namespace{rdf.cpp}::SurfaceType : int { None, Molecule, Residue, Count } |
| Whether to compute RDF wrt. surface of the reference group. | |
Functions | |
| int | gmx::analysismodules::anonymous_namespace{pairdist.cpp}::initSelectionGroups (Selection *sel, const gmx_mtop_t *top, GroupType type) |
| Helper function to initialize the grouping for a selection. | |
| void | gmx::analysismodules::anonymous_namespace{sasa.cpp}::add_rec (t_conect c[], int i, int j, real d2) |
| Updates nearest neighbor information for a surface dot. More... | |
| void | gmx::analysismodules::anonymous_namespace{sasa.cpp}::do_conect (const char *fn, int n, rvec x[]) |
| Adds CONECT records for surface dots. More... | |
| void | gmx::analysismodules::anonymous_namespace{sasa.cpp}::connolly_plot (const char *fn, int ndots, const real dots[], rvec x[], t_atoms *atoms, t_symtab *symtab, PbcType pbcType, const matrix box, gmx_bool bIncludeSolute) |
| Plots the surface into a PDB file, optionally including the original atoms. | |
| void | gmx::analysismodules::anonymous_namespace{sasa.cpp}::computeAreas (const Selection &surfaceSel, const Selection &sel, const std::vector< real > &atomAreas, const std::vector< real > &dgsFactor, real *totalAreaOut, real *dgsolvOut, AnalysisDataHandle atomAreaHandle, AnalysisDataHandle resAreaHandle, std::vector< real > *resAreaWork) |
| Helper method to compute the areas for a single selection. More... | |
| template<class ModuleInfo > | |
| void | gmx::anonymous_namespace{modules.cpp}::registerModule (CommandLineModuleManager *manager, CommandLineModuleGroup group) |
| Convenience method for registering a command-line module for trajectory analysis. More... | |
| void | gmx::registerTrajectoryAnalysisModules (CommandLineModuleManager *manager) |
| Registers all trajectory analysis command-line modules. More... | |
| void | gmx::analysismodules::anonymous_namespace{pairdist.cpp}::PairDistance::initOptions (IOptionsContainer *options, TrajectoryAnalysisSettings *settings) override |
| Initializes options understood by the module. More... | |
| void | gmx::analysismodules::anonymous_namespace{pairdist.cpp}::PairDistance::initAnalysis (const TrajectoryAnalysisSettings &settings, const TopologyInformation &top) override |
| Initializes the analysis. More... | |
| TrajectoryAnalysisModuleDataPointer | gmx::analysismodules::anonymous_namespace{pairdist.cpp}::PairDistance::startFrames (const AnalysisDataParallelOptions &opt, const SelectionCollection &selections) override |
| Starts the analysis of frames. More... | |
| void | gmx::analysismodules::anonymous_namespace{pairdist.cpp}::PairDistance::analyzeFrame (int frnr, const t_trxframe &fr, t_pbc *pbc, TrajectoryAnalysisModuleData *pdata) override |
| Analyzes a single frame. More... | |
| void | gmx::analysismodules::anonymous_namespace{pairdist.cpp}::PairDistance::finishAnalysis (int nframes) override |
| Postprocesses data after frames have been read. More... | |
| void | gmx::analysismodules::anonymous_namespace{pairdist.cpp}::PairDistance::writeOutput () override |
| Writes output into files and/or standard output/error. More... | |
| void | gmx::analysismodules::anonymous_namespace{rdf.cpp}::Rdf::initOptions (IOptionsContainer *options, TrajectoryAnalysisSettings *settings) override |
| Initializes options understood by the module. More... | |
| void | gmx::analysismodules::anonymous_namespace{rdf.cpp}::Rdf::optionsFinished (TrajectoryAnalysisSettings *settings) override |
| Called after all option values have been set. More... | |
| void | gmx::analysismodules::anonymous_namespace{rdf.cpp}::Rdf::initAnalysis (const TrajectoryAnalysisSettings &settings, const TopologyInformation &top) override |
| Initializes the analysis. More... | |
| void | gmx::analysismodules::anonymous_namespace{rdf.cpp}::Rdf::initAfterFirstFrame (const TrajectoryAnalysisSettings &settings, const t_trxframe &fr) override |
| Performs additional initialization after reading the first frame. More... | |
| TrajectoryAnalysisModuleDataPointer | gmx::analysismodules::anonymous_namespace{rdf.cpp}::Rdf::startFrames (const AnalysisDataParallelOptions &opt, const SelectionCollection &selections) override |
| Starts the analysis of frames. More... | |
| void | gmx::analysismodules::anonymous_namespace{rdf.cpp}::Rdf::analyzeFrame (int frnr, const t_trxframe &fr, t_pbc *pbc, TrajectoryAnalysisModuleData *pdata) override |
| Analyzes a single frame. More... | |
| void | gmx::analysismodules::anonymous_namespace{rdf.cpp}::Rdf::finishAnalysis (int nframes) override |
| Postprocesses data after frames have been read. More... | |
| void | gmx::analysismodules::anonymous_namespace{rdf.cpp}::Rdf::writeOutput () override |
| Writes output into files and/or standard output/error. More... | |
| void | gmx::analysismodules::anonymous_namespace{sasa.cpp}::Sasa::initOptions (IOptionsContainer *options, TrajectoryAnalysisSettings *settings) override |
| Initializes options understood by the module. More... | |
| void | gmx::analysismodules::anonymous_namespace{sasa.cpp}::Sasa::initAnalysis (const TrajectoryAnalysisSettings &settings, const TopologyInformation &top) override |
| Initializes the analysis. More... | |
| TrajectoryAnalysisModuleDataPointer | gmx::analysismodules::anonymous_namespace{sasa.cpp}::Sasa::startFrames (const AnalysisDataParallelOptions &opt, const SelectionCollection &selections) override |
| Starts the analysis of frames. More... | |
| void | gmx::analysismodules::anonymous_namespace{sasa.cpp}::Sasa::analyzeFrame (int frnr, const t_trxframe &fr, t_pbc *pbc, TrajectoryAnalysisModuleData *pdata) override |
| Analyzes a single frame. More... | |
| void | gmx::analysismodules::anonymous_namespace{sasa.cpp}::Sasa::finishAnalysis (int nframes) override |
| Postprocesses data after frames have been read. More... | |
| void | gmx::analysismodules::anonymous_namespace{sasa.cpp}::Sasa::writeOutput () override |
| Writes output into files and/or standard output/error. More... | |
Variables | |
|
const EnumerationArray < DistanceType, const char * > | gmx::analysismodules::anonymous_namespace{pairdist.cpp}::c_distanceTypeNames = { { "min", "max" } } |
| Strings corresponding to DistanceType. | |
| const EnumerationArray < GroupType, const char * > | gmx::analysismodules::anonymous_namespace{pairdist.cpp}::c_groupTypeNames |
| Strings corresponding to GroupType. More... | |
| const EnumerationArray < Normalization, const char * > | gmx::analysismodules::anonymous_namespace{rdf.cpp}::c_normalizationNames |
| String values corresponding to Normalization. More... | |
|
const EnumerationArray < SurfaceType, const char * > | gmx::analysismodules::anonymous_namespace{rdf.cpp}::c_surfaceTypeNames = { { "no", "mol", "res" } } |
| String values corresponding to SurfaceType. | |
Directories | |
| directory | trajectoryanalysis |
| Framework for Trajectory Analysis (trajectoryanalysis) | |
| directory | tests |
| Unit tests for Framework for Trajectory Analysis (trajectoryanalysis). | |
Files | |
| file | analysismodule.cpp |
| Implements classes in analysismodule.h. | |
| file | analysissettings.cpp |
| Implements classes in analysissettings.h. | |
| file | analysissettings_impl.h |
| Declares private implementation class for gmx::TrajectoryAnalysisSettings. | |
| file | cmdlinerunner.cpp |
| Implements gmx::TrajectoryAnalysisCommandLineRunner. | |
| file | angle.cpp |
| Implements gmx::analysismodules::Angle. | |
| file | angle.h |
| Declares trajectory analysis module for angle calculations. | |
| file | convert_trj.cpp |
| Implements gmx::analysismodules::ConvertTrj. | |
| file | convert_trj.h |
| Declares trajectory analysis module for trajectory conversion. | |
| file | distance.cpp |
| Implements gmx::analysismodules::Distance. | |
| file | distance.h |
| Declares trajectory analysis module for distance calculations. | |
| file | dssp.cpp |
| Implements gmx::analysismodules::Dssp. | |
| file | dssp.h |
| Declares trajectory analysis module for secondary structure asignment. | |
| file | extract_cluster.cpp |
| Implements gmx::analysismodules::ExtractCluster. | |
| file | extract_cluster.h |
| Declares trajectory analysis module for extracting frames corresponding to clusters. | |
| file | freevolume.cpp |
| Implements gmx::analysismodules::Freevolume. | |
| file | freevolume.h |
| Declares trajectory analysis module for free volume calculations. | |
| file | gyrate.cpp |
| Implements gmx::analysismodules::Gyrate. | |
| file | gyrate.h |
| Declares trajectory analysis module for distance calculations. | |
| file | hbond.cpp |
| Implements gmx::analysismodules::Hbond2. | |
| file | hbond.h |
| Declares trajectory analysis module for computation and analyzation of hydrogen bonds. | |
| file | isotope.cpp |
| Declares helper functions for assigning isotops from atom names. | |
| file | isotope.h |
| Declares helper functions for assigning isotops from atom names. | |
| file | msd.cpp |
| Defines the trajectory analysis module for mean squared displacement calculations. | |
| file | msd.h |
| Declares trajectory analysis module for Mean Squared Displacement calculations. | |
| file | pairdist.cpp |
| Implements gmx::analysismodules::PairDistance. | |
| file | pairdist.h |
| Declares trajectory analysis module for pairwise distance calculations. | |
| file | rdf.cpp |
| Implements gmx::analysismodules::Rdf. | |
| file | rdf.h |
| Declares trajectory analysis module for RDF calculations. | |
| file | sasa.cpp |
| Implements gmx::analysismodules::Sasa. | |
| file | sasa.h |
| Declares trajectory analysis module for surface area calculations. | |
| file | scattering-debye-sans.cpp |
| Implements class for SANS Debye Scattering. | |
| file | scattering-debye-sans.h |
| Declares class for SANS Debye Scattering. | |
| file | scattering-debye-saxs.cpp |
| Implements class for SAXS Debye Scattering. | |
| file | scattering-debye-saxs.h |
| Declares class for SAXS Debye Scattering. | |
| file | scattering-debye.cpp |
| Implementation of base class for Debye Scattering. | |
| file | scattering-debye.h |
| Declares base class for Debye Scattering. | |
| file | scattering.cpp |
| Implements gmx::analysismodules::scattering. | |
| file | scattering.h |
| Declares trajectory analysis module for small angle scattering calculations. | |
| file | scatteringfactors.cpp |
| Implements helper functions for reading structure factors from datafile. | |
| file | scatteringfactors.h |
| Declares helper functions for reading structure factors from datafile. | |
| file | select.cpp |
| Implements gmx::analysismodules::Select. | |
| file | select.h |
| Declares trajectory analysis module for basic selection information. | |
| file | trajectory.cpp |
| Implements gmx::analysismodules::Trajectory. | |
| file | trajectory.h |
| Declares trajectory analysis module for basic trajectory information. | |
| file | unionfind.h |
| Implements gmx::UnionFinder and gmx::MappedUnionFinder. | |
| file | modules.cpp |
| Implements registerTrajectoryAnalysisModules(). | |
| file | modules.h |
| Generic interface for accessing trajectory analysis modules. | |
| file | runnercommon.cpp |
| Implements gmx::TrajectoryAnalysisRunnerCommon. | |
| file | runnercommon.h |
| Declares gmx::TrajectoryAnalysisRunnerCommon. | |
| file | angle.cpp |
| Tests for functionality of the "angle" trajectory analysis module. | |
| file | clustsize.cpp |
| Tests for gmx clustsize. | |
| file | cmdlinerunner.cpp |
| Tests for general functionality in gmx::TrajectoryAnalysisCommandLineRunner. | |
| file | convert_trj.cpp |
| Tests for functionality of the "convert-trj" trajectory analysis module. | |
| file | distance.cpp |
| Tests for functionality of the "distance" trajectory analysis module. | |
| file | dssp.cpp |
| Tests for functionality of the "dssp" trajectory analysis module. | |
| file | extract_cluster.cpp |
| Tests for functionality of the "extract-cluster" trajectory analysis module. | |
| file | freevolume.cpp |
| Tests for functionality of the "angle" trajectory analysis module. | |
| file | gyrate.cpp |
| Tests for functionality of the "gyrate" trajectory analysis module. | |
| file | hbond.cpp |
| Tests for functionality of the "hbond" trajectory analysis module. | |
| file | moduletest.cpp |
| Implements classes in moduletest.h. | |
| file | moduletest.h |
| Declares test fixture for TrajectoryAnalysisModule subclasses. | |
| file | msd.cpp |
| Tests for functionality of the "msd" trajectory analysis module. | |
| file | pairdist.cpp |
| Tests for functionality of the "pairdist" trajectory analysis module. | |
| file | rdf.cpp |
| Tests for functionality of the "rdf" trajectory analysis module. | |
| file | sasa.cpp |
| Tests for functionality of the "sasa" trajectory analysis module. | |
| file | scattering.cpp |
| Tests for scattering trajectory analysis module. | |
| file | select.cpp |
| Tests for functionality of the "select" trajectory analysis module. | |
| file | surfacearea.cpp |
Tests for the surface area calculation used by the sasa analysis module. | |
| file | test_selection.cpp |
| Testing/debugging tool for the selection engine. | |
| file | topologyinformation.cpp |
| Tests for TopologyInformation. | |
| file | trajectory.cpp |
| Tests for functionality of the "trajectory" trajectory analysis module. | |
| file | unionfind.cpp |
| Tests for the union-find implementation in unionfind.h. | |
| file | topologyinformation.cpp |
| Implements classes in topologyinformation.h. | |
| file | analysismodule.h |
| Declares gmx::TrajectoryAnalysisModule and gmx::TrajectoryAnalysisModuleData. | |
| file | analysissettings.h |
| Declares gmx::TrajectoryAnalysisSettings. | |
| file | cmdlinerunner.h |
| Declares gmx::TrajectoryAnalysisCommandLineRunner. | |
| file | topologyinformation.h |
| Declares gmx::TopologyInformation. | |
| void gmx::analysismodules::anonymous_namespace{sasa.cpp}::add_rec | ( | t_conect | c[], |
| int | i, | ||
| int | j, | ||
| real | d2 | ||
| ) |
Updates nearest neighbor information for a surface dot.
| [in,out] | c | Nearest neighbor information array to update. |
| [in] | i | Index in c to update. |
| [in] | j | Index of the other surface dot to add to the array. |
| [in] | d2 | Squared distance between i and j. |
|
overridevirtual |
Analyzes a single frame.
| [in] | frnr | Frame number, a zero-based index that uniquely identifies the frame. |
| [in] | fr | Current frame. |
| [in] | pbc | Periodic boundary conditions for fr. |
| [in,out] | pdata | Data structure for frame-local data. |
This method is called once for each frame to be analyzed, and should analyze the positions provided in the selections. Data handles and selections should be obtained from the pdata structure.
For threaded analysis, this method is called asynchronously in different threads to analyze different frames. The pdata structure is one of the structures created with startFrames(), but no assumptions should be made about which of these data structures is used. It is guaranteed that two instances of analyzeFrame() are not running concurrently with the same pdata data structure. Any access to data structures not stored in pdata should be designed to be thread-safe.
Implements gmx::TrajectoryAnalysisModule.
|
overridevirtual |
Analyzes a single frame.
| [in] | frnr | Frame number, a zero-based index that uniquely identifies the frame. |
| [in] | fr | Current frame. |
| [in] | pbc | Periodic boundary conditions for fr. |
| [in,out] | pdata | Data structure for frame-local data. |
This method is called once for each frame to be analyzed, and should analyze the positions provided in the selections. Data handles and selections should be obtained from the pdata structure.
For threaded analysis, this method is called asynchronously in different threads to analyze different frames. The pdata structure is one of the structures created with startFrames(), but no assumptions should be made about which of these data structures is used. It is guaranteed that two instances of analyzeFrame() are not running concurrently with the same pdata data structure. Any access to data structures not stored in pdata should be designed to be thread-safe.
Implements gmx::TrajectoryAnalysisModule.
|
overridevirtual |
Analyzes a single frame.
| [in] | frnr | Frame number, a zero-based index that uniquely identifies the frame. |
| [in] | fr | Current frame. |
| [in] | pbc | Periodic boundary conditions for fr. |
| [in,out] | pdata | Data structure for frame-local data. |
This method is called once for each frame to be analyzed, and should analyze the positions provided in the selections. Data handles and selections should be obtained from the pdata structure.
For threaded analysis, this method is called asynchronously in different threads to analyze different frames. The pdata structure is one of the structures created with startFrames(), but no assumptions should be made about which of these data structures is used. It is guaranteed that two instances of analyzeFrame() are not running concurrently with the same pdata data structure. Any access to data structures not stored in pdata should be designed to be thread-safe.
Implements gmx::TrajectoryAnalysisModule.
| void gmx::analysismodules::anonymous_namespace{sasa.cpp}::computeAreas | ( | const Selection & | surfaceSel, |
| const Selection & | sel, | ||
| const std::vector< real > & | atomAreas, | ||
| const std::vector< real > & | dgsFactor, | ||
| real * | totalAreaOut, | ||
| real * | dgsolvOut, | ||
| AnalysisDataHandle | atomAreaHandle, | ||
| AnalysisDataHandle | resAreaHandle, | ||
| std::vector< real > * | resAreaWork | ||
| ) |
Helper method to compute the areas for a single selection.
| [in] | surfaceSel | The calculation selection. |
| [in] | sel | The selection to compute the areas for (can be surfaceSel or one of the output selections). |
| [in] | atomAreas | Atom areas for each position in surfaceSel. |
| [in] | dgsFactor | Free energy coefficients for each position in surfaceSel. If empty, free energies are not calculated. |
| [out] | totalAreaOut | Total area of sel (sum of atom areas it selects). |
| [out] | dgsolvOut | Solvation free energy. Will be zero of dgsFactor is empty. |
| atomAreaHandle | Data handle to use for storing atom areas for sel. | |
| resAreaHandle | Data handle to use for storing residue areas for sel. | |
| resAreaWork | Work array for accumulating the residue areas. If empty, atom and residue areas are not calculated. |
atomAreaHandle and resAreaHandle are not used if resAreaWork is empty.
| void gmx::analysismodules::anonymous_namespace{sasa.cpp}::do_conect | ( | const char * | fn, |
| int | n, | ||
| rvec | x[] | ||
| ) |
Adds CONECT records for surface dots.
| [in] | fn | PDB file to append the CONECT records to. |
| [in] | n | Number of dots in x. |
| [in] | x | Array of surface dot positions. |
Adds a CONECT record that connects each surface dot to its two nearest neighbors. The function is copied verbatim from the old gmx_sas.c implementation.
|
overridevirtual |
Postprocesses data after frames have been read.
| [in] | nframes | Total number of frames processed. |
This function is called after all finishFrames() calls have been called. nframes will equal the number of calls to analyzeFrame() that have occurred.
Implements gmx::TrajectoryAnalysisModule.
|
overridevirtual |
Postprocesses data after frames have been read.
| [in] | nframes | Total number of frames processed. |
This function is called after all finishFrames() calls have been called. nframes will equal the number of calls to analyzeFrame() that have occurred.
Implements gmx::TrajectoryAnalysisModule.
|
overridevirtual |
Postprocesses data after frames have been read.
| [in] | nframes | Total number of frames processed. |
This function is called after all finishFrames() calls have been called. nframes will equal the number of calls to analyzeFrame() that have occurred.
Implements gmx::TrajectoryAnalysisModule.
|
overridevirtual |
Performs additional initialization after reading the first frame.
When this function is called, selections are the same as in initAnalysis(), i.e., they have not been evaluated for the first frame.
It is necessary to override this method only if the module needs to do initialization for which it requires data from the first frame.
The default implementation does nothing.
Reimplemented from gmx::TrajectoryAnalysisModule.
|
overridevirtual |
Initializes the analysis.
| [in] | settings | Settings to pass to and from the module. |
| [in] | top | Topology information. |
When this function is called, selections have been initialized based on user input, and a topology has been loaded if provided by the user. For dynamic selections, the selections have been evaluated to the largest possible selection, i.e., the selections passed to analyzeFrame() are always a subset of the selections provided here.
Implements gmx::TrajectoryAnalysisModule.
|
overridevirtual |
Initializes the analysis.
| [in] | settings | Settings to pass to and from the module. |
| [in] | top | Topology information. |
When this function is called, selections have been initialized based on user input, and a topology has been loaded if provided by the user. For dynamic selections, the selections have been evaluated to the largest possible selection, i.e., the selections passed to analyzeFrame() are always a subset of the selections provided here.
Implements gmx::TrajectoryAnalysisModule.
|
overridevirtual |
Initializes the analysis.
| [in] | settings | Settings to pass to and from the module. |
| [in] | top | Topology information. |
When this function is called, selections have been initialized based on user input, and a topology has been loaded if provided by the user. For dynamic selections, the selections have been evaluated to the largest possible selection, i.e., the selections passed to analyzeFrame() are always a subset of the selections provided here.
Implements gmx::TrajectoryAnalysisModule.
|
overridevirtual |
Initializes options understood by the module.
| [in,out] | options | Options object to add the options to. |
| [in,out] | settings | Settings to pass to and from the module. |
This method is called first after the constructor, and it should add options understood by the module to options. Output values from options (including selections) should be stored in member variables.
In addition to initializing the options, this method can also provide information about the module's requirements using the settings object; see TrajectoryAnalysisSettings for more details.
If settings depend on the option values provided by the user, see optionsFinished().
Implements gmx::TrajectoryAnalysisModule.
|
overridevirtual |
Initializes options understood by the module.
| [in,out] | options | Options object to add the options to. |
| [in,out] | settings | Settings to pass to and from the module. |
This method is called first after the constructor, and it should add options understood by the module to options. Output values from options (including selections) should be stored in member variables.
In addition to initializing the options, this method can also provide information about the module's requirements using the settings object; see TrajectoryAnalysisSettings for more details.
If settings depend on the option values provided by the user, see optionsFinished().
Implements gmx::TrajectoryAnalysisModule.
|
overridevirtual |
Initializes options understood by the module.
| [in,out] | options | Options object to add the options to. |
| [in,out] | settings | Settings to pass to and from the module. |
This method is called first after the constructor, and it should add options understood by the module to options. Output values from options (including selections) should be stored in member variables.
In addition to initializing the options, this method can also provide information about the module's requirements using the settings object; see TrajectoryAnalysisSettings for more details.
If settings depend on the option values provided by the user, see optionsFinished().
Implements gmx::TrajectoryAnalysisModule.
|
overridevirtual |
Called after all option values have been set.
| [in,out] | settings | Settings to pass to and from the module. |
This method is called after option values have been assigned (but interactive selection input has not yet been performed).
If the module needs to change settings that affect topology loading (can be done using the settings object) or selection initialization (can be done using SelectionOptionInfo) based on option values, this method has to be overridden.
The default implementation does nothing.
Reimplemented from gmx::TrajectoryAnalysisModule.
| void gmx::anonymous_namespace{modules.cpp}::registerModule | ( | CommandLineModuleManager * | manager, |
| CommandLineModuleGroup | group | ||
| ) |
Convenience method for registering a command-line module for trajectory analysis.
| ModuleInfo | Info about trajectory analysis module to wrap. |
ModuleInfo should have static public members const char name[], const char shortDescription[], and gmx::TrajectoryAnalysisModulePointer create().
| void gmx::registerTrajectoryAnalysisModules | ( | CommandLineModuleManager * | manager | ) |
Registers all trajectory analysis command-line modules.
| [in] | manager | Command-line module manager to receive the modules. |
| std::bad_alloc | if out of memory. |
Registers all trajectory analysis modules declared in the library such that they can be run through manager.
|
overridevirtual |
Starts the analysis of frames.
| [in] | opt | Parallel options |
| [in] | selections | Frame-local selection collection object. |
This function is necessary only for threaded parallelization. It is called once for each thread and should initialize a class that contains any required frame-local data in the returned value. The default implementation creates a basic data structure that holds thread-local data handles for all data objects registered with registerAnalysisDataset(), as well as the thread-local selection collection. These can be accessed in analyzeFrame() using the methods in TrajectoryAnalysisModuleData. If other thread-local data is needed, this function should be overridden and it should create an instance of a class derived from TrajectoryAnalysisModuleData.
Reimplemented from gmx::TrajectoryAnalysisModule.
|
overridevirtual |
Starts the analysis of frames.
| [in] | opt | Parallel options |
| [in] | selections | Frame-local selection collection object. |
This function is necessary only for threaded parallelization. It is called once for each thread and should initialize a class that contains any required frame-local data in the returned value. The default implementation creates a basic data structure that holds thread-local data handles for all data objects registered with registerAnalysisDataset(), as well as the thread-local selection collection. These can be accessed in analyzeFrame() using the methods in TrajectoryAnalysisModuleData. If other thread-local data is needed, this function should be overridden and it should create an instance of a class derived from TrajectoryAnalysisModuleData.
Reimplemented from gmx::TrajectoryAnalysisModule.
|
overridevirtual |
Starts the analysis of frames.
| [in] | opt | Parallel options |
| [in] | selections | Frame-local selection collection object. |
This function is necessary only for threaded parallelization. It is called once for each thread and should initialize a class that contains any required frame-local data in the returned value. The default implementation creates a basic data structure that holds thread-local data handles for all data objects registered with registerAnalysisDataset(), as well as the thread-local selection collection. These can be accessed in analyzeFrame() using the methods in TrajectoryAnalysisModuleData. If other thread-local data is needed, this function should be overridden and it should create an instance of a class derived from TrajectoryAnalysisModuleData.
Reimplemented from gmx::TrajectoryAnalysisModule.
|
overridevirtual |
Writes output into files and/or standard output/error.
All output from the module, excluding data written out for each frame during analyzeFrame(), should be confined into this function. This function is guaranteed to be called only after finishAnalysis().
Implements gmx::TrajectoryAnalysisModule.
|
overridevirtual |
Writes output into files and/or standard output/error.
All output from the module, excluding data written out for each frame during analyzeFrame(), should be confined into this function. This function is guaranteed to be called only after finishAnalysis().
Implements gmx::TrajectoryAnalysisModule.
|
overridevirtual |
Writes output into files and/or standard output/error.
All output from the module, excluding data written out for each frame during analyzeFrame(), should be confined into this function. This function is guaranteed to be called only after finishAnalysis().
Implements gmx::TrajectoryAnalysisModule.
| const EnumerationArray<GroupType, const char*> gmx::analysismodules::anonymous_namespace{pairdist.cpp}::c_groupTypeNames |
Strings corresponding to GroupType.
| const EnumerationArray<Normalization, const char*> gmx::analysismodules::anonymous_namespace{rdf.cpp}::c_normalizationNames |
String values corresponding to Normalization.
 1.8.5
1.8.5