  gmx gmx | Generic GROMACS namespace |
   compat compat | Compatibility aliases for standard library features |
    not_null not_null | Restricts a pointer or smart pointer to only hold non-null values |
   detail detail | |
    PaddingTraits PaddingTraits | Traits classes for handling padding for types used with PaddedVector |
    extents_analyse< R, E0, StaticExtents...> extents_analyse< R, E0, StaticExtents...> | Enable querying extent of specific rank by splitting a static extents off the variadic template arguments |
    extents_analyse< R, dynamic_extent, StaticExtents...> extents_analyse< R, dynamic_extent, StaticExtents...> | Enable querying extent of specific rank by splitting a dynamic extent off the variadic template arguments |
    extents_analyse< 0 > extents_analyse< 0 > | Specialisation for rank 0 extents analysis. Ends recursive rank analysis |
   internal internal | |
    SimdTraits SimdTraits | Simd traits |
   test test | Testing utilities namespace |
    AnalysisDataTestInputPointSet AnalysisDataTestInputPointSet | Represents a single set of points in AnalysisDataTestInputFrame structure |
    AnalysisDataTestInputFrame AnalysisDataTestInputFrame | Represents a single frame in AnalysisDataTestInput structure |
    AnalysisDataTestInput AnalysisDataTestInput | Represents static input data for AbstractAnalysisData tests |
    AnalysisDataTestFixture AnalysisDataTestFixture | Test fixture for AbstractAnalysisData testing |
    ModuleSelection ModuleSelection | Helper class for tests that need an initialized selection |
    ModuleTest ModuleTest | Test fixture to test matching file types for modules |
    SetAtomsSupportedFiles SetAtomsSupportedFiles | Helper to test supported file names |
    SetAtomsUnSupportedFiles SetAtomsUnSupportedFiles | Helper to test supported file names |
    AnyOutputSupportedFiles AnyOutputSupportedFiles | Helper to test supported file names |
    NoOptionalOutput NoOptionalOutput | Helper to test support for disabling all output options |
    OutputSelectorDeathTest OutputSelectorDeathTest | Helper to test that invalid selection is rejected |
    SetVelocitySupportedFiles SetVelocitySupportedFiles | Helper to test supported file names |
    SetVelocityUnSupportedFiles SetVelocityUnSupportedFiles | Helper to test supported file names |
    SetForceSupportedFiles SetForceSupportedFiles | Helper to test supported file names |
    SetForceUnSupportedFiles SetForceUnSupportedFiles | Helper to test supported file names |
    SetPrecisionSupportedFiles SetPrecisionSupportedFiles | Helper to test supported file names |
    SetPrecisionUnSupportedFiles SetPrecisionUnSupportedFiles | Helper to test supported file names |
    FlagTest FlagTest | Test fixture to test user inputs |
    TestSystem TestSystem | Description of the system used for testing |
    AllocatorTest AllocatorTest | Templated test fixture |
    CommandLine CommandLine | Helper class for tests that need to construct command lines |
    CommandLineTestHelper CommandLineTestHelper | Helper class for tests that construct command lines that need to reference existing files |
    CommandLineTestBase CommandLineTestBase | Test fixture for tests that call a single command-line program with input/output files |
    ConfMatch ConfMatch | Match the contents as an gro file |
    IFileMatcher IFileMatcher | Represents a file matcher, matching file contents against reference (or other) data |
    IFileMatcherSettings IFileMatcherSettings | Represents a factory for creating a file matcher |
    TextFileMatch TextFileMatch | Use a ITextBlockMatcher for matching the contents |
    NoContentsMatch NoContentsMatch | Do not check the contents of the file |
    InteractiveTestHelper InteractiveTestHelper | Helper class for testing interactive sessions |
    LoggerTestHelper LoggerTestHelper | Helper class for tests to check output written to a logger |
    TestReferenceData TestReferenceData | Handles creation of and comparison to test reference data |
    TestReferenceChecker TestReferenceChecker | Handles comparison to test reference data |
    StdioTestHelper StdioTestHelper | Helper class for tests where code reads directly from stdin |
    StringTestBase StringTestBase | Test fixture for tests that check string formatting |
    FloatingPointDifference FloatingPointDifference | Computes and represents a floating-point difference value |
    FloatingPointTolerance FloatingPointTolerance | Specifies a floating-point comparison tolerance and checks whether a difference is within the tolerance |
    TestException TestException | Exception class for reporting errors in tests |
    TestFileManager TestFileManager | Helper for tests that need input and output files |
    TestFileInputRedirector TestFileInputRedirector | In-memory implementation for IFileInputRedirector for tests |
    TestFileOutputRedirector TestFileOutputRedirector | In-memory implementation of IFileOutputRedirector for tests |
    TestOptionsProvider TestOptionsProvider | Provides additional options for the test executable |
    ITextBlockMatcher ITextBlockMatcher | Represents a text matcher, matching text stream contents against reference data |
    ITextBlockMatcherSettings ITextBlockMatcherSettings | Represents a factory for creating a text matcher |
    ExactTextMatch ExactTextMatch | Use an exact text match (the contents should be exactly equal) |
    NoTextMatch NoTextMatch | Do not match the text (the contents are ignored) |
    FilteringExactTextMatch FilteringExactTextMatch | Use an exact text match after scrubbing lines of the text that match the supplied regular expressions |
    TprAndFileManager TprAndFileManager | Helper to bundle generated TPR and the file manager to clean it up |
    XvgMatch XvgMatch | Match the contents as an xvg file |
   AnalysisDataModuleManager AnalysisDataModuleManager | Encapsulates handling of data modules attached to AbstractAnalysisData |
   AnalysisDataStorageFrame AnalysisDataStorageFrame | Allows assigning values for a data frame in AnalysisDataStorage |
   AnalysisDataStorage AnalysisDataStorage | Helper class that implements storage of data |
   ArrayRef ArrayRef | STL-like interface to a C array of T (or part of a std container of T) |
   AnalysisDataParallelOptions AnalysisDataParallelOptions | Parallelization options for analysis data objects |
   Awh Awh | Coupling of the accelerated weight histogram method (AWH) with the system |
   DensityFittingModuleInfo DensityFittingModuleInfo | Information about the density fitting module |
   CommandLineHelpContext CommandLineHelpContext | Context information for writing out command-line help |
   GlobalCommandLineHelpContext GlobalCommandLineHelpContext | Helper for passing CommandLineHelpContext into parse_common_args() |
   CommandLineHelpWriter CommandLineHelpWriter | Writes help information for Options |
   CommandLineModuleSettings CommandLineModuleSettings | Settings to pass information between a module and the general runner |
   ICommandLineModule ICommandLineModule | Module that can be run from command line using CommandLineModuleManager |
   CommandLineModuleManager CommandLineModuleManager | Implements a wrapper command-line interface for multiple modules |
   CommandLineModuleGroup CommandLineModuleGroup | Handle to add content to a group added with CommandLineModuleManager::addModuleGroup() |
   ICommandLineOptionsModuleSettings ICommandLineOptionsModuleSettings | Settings to pass information between a CommandLineOptionsModule and generic code that runs it |
   ICommandLineOptionsModule ICommandLineOptionsModule | Module that can be run from a command line and uses gmx::Options for argument processing |
   CommandLineParser CommandLineParser | Implements command-line parsing for Options objects |
   IExecutableEnvironment IExecutableEnvironment | Allows customization of the way various directories are found by CommandLineProgramContext |
   CommandLineProgramContext CommandLineProgramContext | Program context implementation for command line programs |
   TrajectoryFileOpener TrajectoryFileOpener | Low level method to take care of only file opening and closing |
   TrajectoryFrameWriter TrajectoryFrameWriter | Writes coordinate frames to a sink, e.g. a trajectory file |
   ProcessFrameConversion ProcessFrameConversion | ProcessFrameConversion class for handling the running of several analysis steps |
   IFrameConverter IFrameConverter | IFrameConverter interface for manipulating coordinate information |
   IOutputAdapter IOutputAdapter | OutputAdapter class for handling trajectory file flag setting and processing |
   OutputAdapterContainer OutputAdapterContainer | Storage for output adapters that modify the state of a t_trxframe object |
   OutputSelector OutputSelector | OutputSelector class controls setting which coordinates are actually written |
   SetAtoms SetAtoms | SetAtoms class controls availability of atoms data |
   SetBox SetBox | Allows changing box information when writing a coordinate file |
   SetForces SetForces | SetForces class allows changing writing of forces to file |
   SetPrecision SetPrecision | SetPrecision class allows changing file writing precision |
   SetStartTime SetStartTime | SetStartTime class allows changing trajectory time information |
   SetTimeStep SetTimeStep | SetTimeStep class allows changing trajectory time information |
   SetVelocities SetVelocities | SetVelocities class allows changing writing of velocities to file |
   OutputRequirementOptionDirector OutputRequirementOptionDirector | Container for the user input values that will be used by the builder to determine which OutputAdapters should/could/will be registered to the coordinate file writer |
   OutputRequirements OutputRequirements | Finalized version of requirements after processing |
   DomainDecompositionBuilder DomainDecompositionBuilder | Builds a domain decomposition management object |
   FixedCapacityVector FixedCapacityVector | Vector that behaves likes std::vector but has fixed capacity |
   HashedMap HashedMap | Unordered key to value mapping |
   GpuHaloExchange GpuHaloExchange | Manages GPU Halo Exchange object |
   LocalAtomSet LocalAtomSet | A local atom set collects local, global and collective indices of the home atoms on a rank. The indices of the home atoms are automatically updated during domain decomposition, thus gmx::LocalAtomSet::localIndex enables iteration over local atoms properties like coordinates or forces. TODO: add a LocalAtomSet iterator |
   LocalAtomSetManager LocalAtomSetManager | Hands out handles to local atom set indices and triggers index recalculation for all sets upon domain decomposition if run in parallel |
   LocalTopologyChecker LocalTopologyChecker | Has responsibility for checking that the local topology distributed across domains describes a total number of bonded interactions that matches the system topology |
   DomdecOptions DomdecOptions | Structure containing all (command line) options for the domain decomposition |
   EnergyAnalysisFrame EnergyAnalysisFrame | Class describing an energy frame, that is the all the data stored for one energy term at one time step in an energy file |
   EnergyTerm EnergyTerm | Class describing the whole time series of an energy term |
   EnergyNameUnit EnergyNameUnit | Convenience structure for keeping energy name and unit together |
   IEnergyAnalysis IEnergyAnalysis | Interface class overloaded by the separate energy modules |
   SeparatePmeRanksPermitted SeparatePmeRanksPermitted | Class for managing usage of separate PME-only ranks |
   PmeForceSenderGpu PmeForceSenderGpu | Manages sending forces from PME-only ranks to their PP ranks |
   PmePpCommGpu PmePpCommGpu | Manages communication related to GPU buffers between this PME rank and its PP rank |
   MDModulesCheckpointReadingDataOnMain MDModulesCheckpointReadingDataOnMain | Provides the MDModules with the checkpointed data on the main rank |
   MDModulesCheckpointReadingBroadcast MDModulesCheckpointReadingBroadcast | Provides the MDModules with the communication record to broadcast |
   MDModulesWriteCheckpointData MDModulesWriteCheckpointData | Writing the MDModules data to a checkpoint file |
   MrcDensityMapOfFloatReader MrcDensityMapOfFloatReader | Read an mrc/ccp4 file that contains float values |
   MrcDensityMapOfFloatFromFileReader MrcDensityMapOfFloatFromFileReader | Read an mrc density map from a given file |
   MrcDensityMapOfFloatWriter MrcDensityMapOfFloatWriter | Write an mrc/ccp4 file that contains float values |
   MrcDataStatistics MrcDataStatistics | Statistics about mrc data arrays |
   MrcDensitySkewData MrcDensitySkewData | Skew matrix and translation. As named in "EMDB Map Distribution Format Description Version 1.01 (c) emdatabank.org 2014" |
   CrystallographicLabels CrystallographicLabels | Crystallographic labels for mrc data |
   MrcDensityMapHeader MrcDensityMapHeader | A container for the data in mrc density map file formats |
   ArrayRefWithPadding ArrayRefWithPadding | Interface to a C array of T (or part of a std container of T), that includes padding that is suitable for the kinds of SIMD operations GROMACS uses |
   ListOfLists ListOfLists | A list of lists, optimized for performance |
   ClfftInitializer ClfftInitializer | Handle clFFT library init and tear down in RAII style also with mutual exclusion |
   DeviceStreamManager DeviceStreamManager | Device stream and context manager |
   HostAllocationPolicy HostAllocationPolicy | Policy class for configuring gmx::Allocator, to manage allocations of memory that may be needed for e.g. GPU transfers |
   OpenClTraits OpenClTraits | Stub for OpenCL type traits |
   OpenClTraitsBase OpenClTraitsBase | Implements common trait infrastructure for OpenCL types |
   OpenClTraits< cl_context > OpenClTraits< cl_context > | Implements traits for cl_context |
   OpenClTraits< cl_command_queue > OpenClTraits< cl_command_queue > | Implements traits for cl_command_queue |
   OpenClTraits< cl_program > OpenClTraits< cl_program > | Implements traits for cl_program |
   OpenClTraits< cl_kernel > OpenClTraits< cl_kernel > | Implements traits for cl_kernel |
   ClHandle ClHandle | Wrapper of OpenCL type cl_type to implement RAII |
   CpuInfo CpuInfo | Detect CPU capabilities and basic logical processor info |
    LogicalProcessor LogicalProcessor | Entry with basic information for a single processing unit |
   HardwareTopology HardwareTopology | Information about packages, cores, processing units, numa, caches |
    Cache Cache | Information about a single cache level |
    Core Core | Information about a single core in a package |
    Device Device | Information about a single PCI device |
    LogicalProcessor LogicalProcessor | Information about package, core and processing unit for a logical processor |
    Machine Machine | Hardware topology information about the entire machine |
    Numa Numa | Information about all numa nodes |
    NumaNode NumaNode | Information about each numa node in system |
    Package Package | Information about a single package in the system |
    ProcessingUnit ProcessingUnit | Information about a single processing unit (in a core) |
   EnumerationArray EnumerationArray | Wrapper for a C-style array with size and indexing defined by an enum. Useful for declaring arrays of enum names for debug or other printing. An ArrayRef<DataType> may be constructed from an object of this type |
   TranslateAndScale TranslateAndScale | Transform coordinates in three dimensions by first translating, then scaling them |
   AffineTransformation AffineTransformation | Affine transformation of three-dimensional coordinates |
   DensitySimilarityMeasure DensitySimilarityMeasure | Measure similarity and gradient between densities |
   DensityFittingForce DensityFittingForce | Manages evaluation of density-fitting forces for particles that were spread with a kernel |
   ExponentialMovingAverageState ExponentialMovingAverageState | Store the state of exponential moving averages |
   ExponentialMovingAverage ExponentialMovingAverage | Evaluate the exponential moving average with bias correction |
   GaussianSpreadKernelParameters GaussianSpreadKernelParameters | Parameters for density spreading kernels |
    PositionAndAmplitude PositionAndAmplitude | Parameters that describe the kernel position and amplitude |
    Shape Shape | Shape parameters for Gaussian spreading kernels describe the kernel shape |
   GaussTransform3D GaussTransform3D | Sums Gaussian values at three dimensional lattice coordinates. The Gaussian is defined as 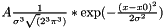 |
   MultiDimArray MultiDimArray | Multidimensional array that manages its own memory |
   PaddedVector PaddedVector | PaddedVector is a container of elements in contiguous storage that allocates extra memory for safe SIMD-style loads for operations used in GROMACS |
   CheckpointHandler CheckpointHandler | Class handling the checkpoint signal |
   Constraints Constraints | Handles constraints |
   AtomPair AtomPair | A pair of atoms indexes |
   MDAtoms MDAtoms | Contains a C-style t_mdatoms while managing some of its memory with C++ vectors with allocators |
   ResetHandler ResetHandler | Class handling the reset of counters |
   SettleData SettleData | Data for executing SETTLE constraining |
   shakedata shakedata | Working data for the SHAKE algorithm |
   SimulationSignal SimulationSignal | POD-style object used by mdrun ranks to set and receive signals within and between simulations |
   SimulationSignaller SimulationSignaller | Object used by mdrun ranks to signal to each other at this step |
   StopHandler StopHandler | Class handling the stop signal |
   StopConditionSignal StopConditionSignal | Class setting the stop signal based on gmx_get_stop_condition() |
   StopConditionTime StopConditionTime | Class setting the stop signal based on maximal run time |
   StopHandlerBuilder StopHandlerBuilder | Class preparing the creation of a StopHandler |
   Update Update | Contains data for update phase |
   UpdateGroups UpdateGroups | Owns the update grouping and related data |
   UpdateGroupsCog UpdateGroupsCog | Class for managing and computing centers of geometry of update groups |
   VirtualSitesHandler VirtualSitesHandler | Class that handles construction of vsites and spreading of vsite forces |
   WholeMoleculeTransform WholeMoleculeTransform | This class manages a coordinate buffer with molecules not split over periodic boundary conditions for use in force calculations which require whole molecules |
   LegacyMdrunOptions LegacyMdrunOptions | This class provides the same command-line option functionality to both CLI and API sessions |
   MDModules MDModules | Manages the collection of all modules used for mdrun |
   MembedHolder MembedHolder | Membed SimulatorBuilder parameter type |
   Mdrunner Mdrunner | Runner object for supporting setup and execution of mdrun |
   MdrunnerBuilder MdrunnerBuilder | Build a gmx::Mdrunner |
   SimulationContext SimulationContext | Simulation environment and configuration |
   SimulatorConfig SimulatorConfig | Simulation configuation settings |
   SimulatorStateData SimulatorStateData | Data for a specific simulation state |
   SimulatorEnv SimulatorEnv | Collection of environmental information for a simulation |
   Profiling Profiling | Collection of profiling information |
   ConstraintsParam ConstraintsParam | Collection of constraint parameters |
   LegacyInput LegacyInput | Collection of legacy input information |
   InteractiveMD InteractiveMD | SimulatorBuilder parameter type for InteractiveMD |
   IonSwapping IonSwapping | Parameter type for IonSwapping SimulatorBuilder component |
   TopologyData TopologyData | Collection of handles to topology information |
   BoxDeformationHandle BoxDeformationHandle | Handle to information about the box |
   SimulatorBuilder SimulatorBuilder | Class preparing the creation of Simulator objects |
   MDModulesNotifier MDModulesNotifier | Organizes notifications about an event of interest to modules |
   BuildMDModulesNotifier< CurrentCallParameter, CallParameter...> BuildMDModulesNotifier< CurrentCallParameter, CallParameter...> | Template specialization to assemble MDModulesNotifier |
   MDModulesAtomsRedistributedSignal MDModulesAtomsRedistributedSignal | Notification that atoms may have been redistributed |
   MDModulesEnergyOutputToDensityFittingRequestChecker MDModulesEnergyOutputToDensityFittingRequestChecker | Check if module outputs energy to a specific field |
   MDModulesEnergyOutputToQMMMRequestChecker MDModulesEnergyOutputToQMMMRequestChecker | Check if QMMM module outputs energy to a specific field |
   EnergyCalculationFrequencyErrors EnergyCalculationFrequencyErrors | Collect errors for the energy calculation frequency |
   SimulationTimeStep SimulationTimeStep | Provides the simulation time step in ps |
   CoordinatesAndBoxPreprocessed CoordinatesAndBoxPreprocessed | Provides coordinates and simulation box |
   MdRunInputFilename MdRunInputFilename | Mdrun input filename |
   EdrOutputFilename EdrOutputFilename | Energy trajectory output filename from Mdrun |
   QMInputFileName QMInputFileName | Notification for QM program input filename provided by user as command-line argument for grompp |
   EnsembleTemperature EnsembleTemperature | Provides the constant ensemble temperature |
   MDModulesNotifiers MDModulesNotifiers | Group of notifers to organize that MDModules can receive callbacks they subscribe to |
   accessor_basic accessor_basic | The most basic memory access model for mdspan |
   extents extents | Multidimensional extents with static and dynamic dimensions |
   layout_right layout_right | Right-aligned array layout indexer. Carries the mapping class performing the translation from multidimensional index to one-dimensional number |
    mapping mapping | Mapping from multidimensional indices within extents to 1D index |
   basic_mdspan basic_mdspan | Multidimensional array indexing and memory access with flexible mapping and access model |
   CheckpointData CheckpointData | } |
   IsSerializableEnum IsSerializableEnum | { |
   ReadCheckpointDataHolder ReadCheckpointDataHolder | Holder for read checkpoint data |
   WriteCheckpointDataHolder WriteCheckpointDataHolder | Holder for write checkpoint data |
   ForceBuffersView ForceBuffersView | A view of the force buffer |
   ForceBuffers ForceBuffers | Object that holds the force buffers |
   ForceWithShiftForces ForceWithShiftForces | Container for force and virial for algorithms that compute shift forces for virial calculation |
   ForceWithVirial ForceWithVirial | Container for force and virial for algorithms that provide their own virial tensor contribution |
   ForceOutputs ForceOutputs | Force and virial output buffers for use in force computation |
   ForceProviderInput ForceProviderInput | Helper struct that bundles data for passing it over to the force providers |
   ForceProviderOutput ForceProviderOutput | Helper struct bundling the output data of a force provider |
   IForceProvider IForceProvider | Interface for a component that provides forces during MD |
   ForceProviders ForceProviders | Evaluates forces from a collection of gmx::IForceProvider |
   IMDModule IMDModule | Extension module for GROMACS simulations |
   IMDOutputProvider IMDOutputProvider | Interface for handling additional output files during a simulation |
   IMdpOptionProvider IMdpOptionProvider | Interface for handling mdp/tpr input to a mdrun module |
   MtsLevel MtsLevel | Setting for a single level for multiple time step integration |
   GromppMtsOpts GromppMtsOpts | Struct for passing the MTS mdp options to setupMtsLevels() |
   ObservablesReducerBuilder ObservablesReducerBuilder | Builder for ObservablesReducer |
   ObservablesReducer ObservablesReducer | Manage reduction of observables for registered subscribers |
   StepWorkload StepWorkload | Describes work done on this domain by the current rank that may change per-step |
   DomainLifetimeWorkload DomainLifetimeWorkload | Describes work done on this domain on every step of its lifetime, but which might change after the next domain partitioning |
   SimulationWorkload SimulationWorkload | Manage what computation is required during the simulation |
   ModularSimulator ModularSimulator | The modular simulator |
   ElementNotFoundError ElementNotFoundError | Exception class signalling that a requested element was not found |
   MissingElementConnectionError MissingElementConnectionError | Exception class signalling that elements were not connected properly |
   SimulationAlgorithmSetupError SimulationAlgorithmSetupError | Exception class signalling that the ModularSimulatorAlgorithm was set up in an incompatible way |
   CheckpointError CheckpointError | Exception class signalling an error in reading or writing modular checkpoints |
   EnergyAccumulator EnergyAccumulator | Base energy accumulator class, only specializations are used |
   BenchmarkSystem BenchmarkSystem | Description of the system used for benchmarking |
   Range Range | Defines a range of integer numbers and accompanying operations |
    iterator iterator | An iterator that loops over a range of integers |
   CoulombCalculator CoulombCalculator | Base Coulomb calculator class, only specializations are used |
   CoulombCalculator< KernelCoulombType::RF > CoulombCalculator< KernelCoulombType::RF > | Specialized calculator for RF |
   CoulombCalculator< KernelCoulombType::EwaldAnalytical > CoulombCalculator< KernelCoulombType::EwaldAnalytical > | Specialized calculator for Ewald using an analytic approximation |
   CoulombCalculator< KernelCoulombType::EwaldTabulated > CoulombCalculator< KernelCoulombType::EwaldTabulated > | Specialized calculator for Ewald using tabulated functions |
   DiagonalMasker DiagonalMasker | Base Coulomb calculator class, only specializations are used |
   DiagonalMasker< nR, kernelLayout, KernelLayoutClusterRatio::JSizeEqualsISize > DiagonalMasker< nR, kernelLayout, KernelLayoutClusterRatio::JSizeEqualsISize > | Specialized masker for JSizeEqualsISize |
   DiagonalMasker< nR, kernelLayout, KernelLayoutClusterRatio::JSizeIsDoubleISize > DiagonalMasker< nR, kernelLayout, KernelLayoutClusterRatio::JSizeIsDoubleISize > | Specialized masker for JSizeIsDoubleISize |
   DiagonalMasker< nR, kernelLayout, KernelLayoutClusterRatio::JSizeIsHalfISize > DiagonalMasker< nR, kernelLayout, KernelLayoutClusterRatio::JSizeIsHalfISize > | Specialized masker for JSizeIsHalfISize |
   EnergyAccumulator< useEnergyGroups, false > EnergyAccumulator< useEnergyGroups, false > | Specialized energy accumulator class for no energy calculation |
   EnergyAccumulator< false, true > EnergyAccumulator< false, true > | Specialized energy accumulator class for energy accumulation without energy groups |
   EnergyAccumulator< true, true > EnergyAccumulator< true, true > | Specialized energy accumulator class for energy accumulation with energy groups |
   EnergyGroupsPerCluster EnergyGroupsPerCluster | Holds energy group indices for use in EnergyAccumulator<true, true> |
   LennardJonesCalculator LennardJonesCalculator | Base LJ calculator class, only specializations are used |
   LennardJonesCalculator< false, InteractionModifiers::PotShift > LennardJonesCalculator< false, InteractionModifiers::PotShift > | Specialized calculator for LJ with potential shift and no energy calculation |
   LennardJonesCalculator< true, InteractionModifiers::PotShift > LennardJonesCalculator< true, InteractionModifiers::PotShift > | Specialized calculator for LJ with potential shift and energy calculation |
   LennardJonesCalculator< calculateEnergies, InteractionModifiers::ForceSwitch > LennardJonesCalculator< calculateEnergies, InteractionModifiers::ForceSwitch > | Specialized calculator for LJ with force switch |
   LennardJonesCalculator< calculateEnergies, InteractionModifiers::PotSwitch > LennardJonesCalculator< calculateEnergies, InteractionModifiers::PotSwitch > | Specialized calculator for LJ with potential switch |
   TextTableFormatter TextTableFormatter | Formats rows of a table for text output |
   HelpManager HelpManager | Helper for providing interactive online help |
   AbstractSimpleHelpTopic AbstractSimpleHelpTopic | Abstract base class for help topics that have simple text and no subtopics |
   AbstractCompositeHelpTopic AbstractCompositeHelpTopic | Abstract base class for help topics that have simple text and subtopics |
   SimpleHelpTopic SimpleHelpTopic | Template for simple implementation of AbstractSimpleHelpTopic |
   CompositeHelpTopic CompositeHelpTopic | Template for simple implementation of AbstractCompositeHelpTopic |
   HelpLinks HelpLinks | Hyperlink data for writing out help |
   HelpWriterContext HelpWriterContext | Context information for writing out help |
   IHelpTopic IHelpTopic | Provides a single online help topic |
   AbstractOptionStorage AbstractOptionStorage | Abstract base class for converting, validating, and storing option values |
   AbstractOptionSection AbstractOptionSection | Base class for specifying option section properties |
   AbstractOptionSectionHandle AbstractOptionSectionHandle | Base class for handles to option sections |
   OptionsBehaviorCollection OptionsBehaviorCollection | Container for IOptionsBehavior objects |
   OptionManagerContainer OptionManagerContainer | Container to keep managers added with Options::addManager() and pass them to options |
   OptionsAssigner OptionsAssigner | Decorator class for assigning values to Options |
   OptionSection OptionSection | Declares a simple option section |
   OptionSectionHandle OptionSectionHandle | Allows adding options to an OptionSection |
   OptionStorageTemplate OptionStorageTemplate | Templated base class for constructing option value storage classes |
   OptionStorageTemplateSimple OptionStorageTemplateSimple | Simplified option storage template for options that have one-to-one value conversion |
   OptionsVisitor OptionsVisitor | Pure interface for visiting options in a Options object |
   OptionsTypeVisitor OptionsTypeVisitor | Abstract base class for visiting options of a particular type |
   OptionsIterator OptionsIterator | Decorator class for visiting options in a Options object |
   OptionsModifyingVisitor OptionsModifyingVisitor | Pure interface for visiting options in a Options object, allowing modifications |
   OptionsModifyingTypeVisitor OptionsModifyingTypeVisitor | Abstract base class for visiting options of a particular type, allowing modifications |
   OptionsModifyingIterator OptionsModifyingIterator | Decorator class for visiting options in a Options object, allowing changes |
   RepeatingOptionSectionHandle RepeatingOptionSectionHandle | Allows adding options to an RepeatingOptionSection |
   RepeatingOptionSection RepeatingOptionSection | Declares an option section that creates a structure for each instance |
   OptionValueConverterSimple OptionValueConverterSimple | Helper for converting from Any to a given type |
   PullCoordExpressionParser PullCoordExpressionParser | Class with a mathematical expression and parser |
   ExponentialDistribution ExponentialDistribution | Exponential distribution |
    param_type param_type | Exponential distribution parameters |
   GammaDistribution GammaDistribution | Gamma distribution |
    param_type param_type | Gamma distribution parameters |
   NormalDistribution NormalDistribution | Normal distribution |
    param_type param_type | Normal distribution parameters |
   TabulatedNormalDistribution TabulatedNormalDistribution | Tabulated normal random distribution |
    param_type param_type | Normal distribution parameter class (mean and stddev) |
   ThreeFry2x64General ThreeFry2x64General | General implementation class for ThreeFry counter-based random engines |
   ThreeFry2x64 ThreeFry2x64 | ThreeFry2x64 random engine with 20 iteractions |
   ThreeFry2x64Fast ThreeFry2x64Fast | ThreeFry2x64 random engine with 13 iteractions |
   UniformIntDistribution UniformIntDistribution | Uniform integer distribution |
    param_type param_type | Uniform int distribution parameters |
   UniformRealDistribution UniformRealDistribution | Uniform real distribution |
    param_type param_type | Uniform real distribution parameters |
   RestraintManager RestraintManager | Manage the Restraint potentials available for Molecular Dynamics |
   RestraintMDModule RestraintMDModule | MDModule wrapper for Restraint implementations |
   Site Site | Abstraction for a restraint interaction site |
   SelectionFileOption SelectionFileOption | Specifies a special option that provides selections from a file |
   SelectionFileOptionInfo SelectionFileOptionInfo | Wrapper class for accessing and modifying selection file option information |
   Simd4Double Simd4Double | SIMD4 double type |
   Simd4DBool Simd4DBool | SIMD4 variable type to use for logical comparisons on doubles |
   Simd4Float Simd4Float | SIMD4 float type |
   Simd4FBool Simd4FBool | SIMD4 variable type to use for logical comparisons on floats |
   SimdDouble SimdDouble | Double SIMD variable. Available if GMX_SIMD_HAVE_DOUBLE is 1 |
   SimdDInt32 SimdDInt32 | Integer SIMD variable type to use for conversions to/from double |
   SimdDBool SimdDBool | Boolean type for double SIMD data |
   SimdDIBool SimdDIBool | Boolean type for integer datatypes corresponding to double SIMD |
   SimdFloat SimdFloat | Float SIMD variable. Available if GMX_SIMD_HAVE_FLOAT is 1 |
   SimdFInt32 SimdFInt32 | Integer SIMD variable type to use for conversions to/from float |
   SimdFBool SimdFBool | Boolean type for float SIMD data |
   SimdFIBool SimdFIBool | Boolean type for integer datatypes corresponding to float SIMD |
   SimdFloatTag SimdFloatTag | Tag type to select to load SimdFloat with simdLoad(U) |
   SimdDoubleTag SimdDoubleTag | Tag type to select to load SimdDouble with simdLoad(U) |
   SimdFInt32Tag SimdFInt32Tag | Tag type to select to load SimdFInt32 with simdLoad(U) |
   SimdDInt32Tag SimdDInt32Tag | Tag type to select to load SimdDInt32 with simdLoad(U) |
   AlignedArray< float, N > AlignedArray< float, N > | Identical to std::array with GMX_SIMD_FLOAT_WIDTH alignment. Should not be deleted through base pointer (destructor is non-virtual) |
   AlignedArray< double, N > AlignedArray< double, N > | Identical to std::array with GMX_SIMD_DOUBLE_WIDTH alignment. Should not be deleted through base pointer (destructor is non-virtual) |
   SimdSetZeroProxy SimdSetZeroProxy | Proxy object to enable setZero() for SIMD and real types |
   CubicSplineTable CubicSplineTable | Cubic spline interpolation table |
   QuadraticSplineTable QuadraticSplineTable | Quadratic spline interpolation table |
   AnalyticalSplineTableInput AnalyticalSplineTableInput | Specification for analytical table function (name, function, derivative) |
   NumericalSplineTableInput NumericalSplineTableInput | Specification for vector table function (name, function, derivative, spacing) |
   DevelopmentFeatureFlags DevelopmentFeatureFlags | Structure that holds boolean flags corresponding to the development features present enabled through environment variables |
   GpuTaskMapping GpuTaskMapping | Specifies the GPU deviceID_ available for task_ to use |
   GpuTaskAssignmentsBuilder GpuTaskAssignmentsBuilder | Builder for the GpuTaskAssignments for all ranks on this node |
   GpuTaskAssignments GpuTaskAssignments | Contains the GPU task assignment for all ranks on this physical node |
   ConvertTprInfo ConvertTprInfo | Declares gmx convert-tpr |
   ExclusionBlock ExclusionBlock | Describes exclusions for a single atom |
   UnionFinder UnionFinder | Union-find data structure for keeping track of disjoint sets |
   MappedUnionFinder MappedUnionFinder | Extension of UnionFind that supports non-consecutive integer indices as items |
   AlignedAllocationPolicy AlignedAllocationPolicy | Policy class for configuring gmx::Allocator, to manage allocations of aligned memory for SIMD code |
   PageAlignedAllocationPolicy PageAlignedAllocationPolicy | Policy class for configuring gmx::Allocator, to manage allocations of page-aligned memory that can be locked for asynchronous transfer to GPU devices |
   Allocator Allocator | Policy-based memory allocator |
   Any Any | Represents a dynamically typed value of an arbitrary type - deprecated |
   BinaryInformationSettings BinaryInformationSettings | Settings for printBinaryInformation() |
   BoolType BoolType | A clone of a bool as a workaround on the template specialization of std::vector<bool> that is incompatible with ArrayRef |
   DataFileOptions DataFileOptions | Search parameters for DataFileFinder |
   DataFileInfo DataFileInfo | Information about a data file found by DataFileFinder::enumerateFiles() |
   DataFileFinder DataFileFinder | Searches data files from a set of paths |
   DefaultInitializationAllocator DefaultInitializationAllocator | Allocator adaptor that interposes construct() calls to convert value initialization into default initialization |
   DirectoryEnumerator DirectoryEnumerator | Lists files in a directory |
   IFileInputRedirector IFileInputRedirector | Allows overriding file existence checks from code that supports it |
   IFileOutputRedirector IFileOutputRedirector | Allows capturing stdout and file output from code that supports it |
   StandardInputStream StandardInputStream | Text input stream implementation for reading from stdin |
   TextInputFile TextInputFile | Text input stream implementation for reading from a file |
   TextOutputFile TextOutputFile | Text output stream implementation for writing to a file |
   ISerializer ISerializer | Interface for types that convert standard data types into a form suitable for storage or transfer |
   KeyValueTreePath KeyValueTreePath | Identifies an entry in a key-value tree |
   KeyValueTreeBuilder KeyValueTreeBuilder | Root builder for creating trees that have an object at the root |
   KeyValueTreeValueBuilder KeyValueTreeValueBuilder | Builder for KeyValueTreeValue objects |
   KeyValueTreeUniformArrayBuilder KeyValueTreeUniformArrayBuilder | Builder for KeyValueTreeArray objects where all elements are of type T |
   KeyValueTreeObjectArrayBuilder KeyValueTreeObjectArrayBuilder | Builder for KeyValueTreeArray objects where all elements are KeyValueTreeObject objects |
   KeyValueTreeObjectBuilder KeyValueTreeObjectBuilder | Builder for KeyValueTreeObject objects |
   IKeyValueTreeTransformRules IKeyValueTreeTransformRules | Interface to declare rules for transforming key-value trees |
   KeyValueTreeTransformRulesScoped KeyValueTreeTransformRulesScoped | Helper object returned from IKeyValueTreeTransformRules::scopedTransform() |
   KeyValueTreeTransformRuleBuilder KeyValueTreeTransformRuleBuilder | Provides methods to specify one transformation rule |
    AfterFrom AfterFrom | Properties that can be specified after from() |
    ToObject ToObject | Properties that can be specified after from().toObject() |
    ToValue ToValue | Properties that can be specified after from().to() |
   ILogTarget ILogTarget | Target where log output can be written |
   LogEntryWriter LogEntryWriter | Helper class for creating log entries with GMX_LOG |
   LogLevelHelper LogLevelHelper | Represents a single logging level |
   MDLogger MDLogger | Declares a logging interface |
   LoggerBuilder LoggerBuilder | Initializes loggers |
   LoggerOwner LoggerOwner | Manages memory for a logger built with LoggerBuilder |
   MessageStringCollector MessageStringCollector | Helper class for collecting message strings, optionally with context |
   MessageStringContext MessageStringContext | Convenience class for creating a message context |
   PhysicalNodeCommunicator PhysicalNodeCommunicator | Holds a communicator for the physical node of this rank |
   StringCompare StringCompare | Compare object for std::string STL containers and algorithms that supports run-time decision on how to compare |
   StringOutputStream StringOutputStream | Text output stream implementation for writing to an in-memory string |
   StringInputStream StringInputStream | Helper class to convert static string data to a stream |
   StringToEnumValueConverter StringToEnumValueConverter | A class to convert a string to an enum value of type EnumType |
   TextReader TextReader | Reads text from a TextInputStream |
   TextInputStream TextInputStream | Interface for reading text |
   TextOutputStream TextOutputStream | Interface for writing text |
   TextWriter TextWriter | Writes text into a TextOutputStream |
   isIntegralConstant isIntegralConstant | Is true if type is a std::integral_constant |
   no_delete no_delete | Deleter for std::shared_ptr that does nothing |
   NonbondedBenchmarkInfo NonbondedBenchmarkInfo | Declares gmx nonbonded-bench |
   BasicVector BasicVector | C++ class for 3D vectors |
   AbstractAnalysisData AbstractAnalysisData | Abstract base class for all objects that provide data |
   AnalysisData AnalysisData | Parallelizable data container for raw data |
   AnalysisDataHandle AnalysisDataHandle | Handle for inserting data into AnalysisData |
   AbstractAnalysisArrayData AbstractAnalysisArrayData | Abstract base class for data objects that present in-memory data |
   AnalysisArrayData AnalysisArrayData | Simple in-memory data array |
   AnalysisDataValue AnalysisDataValue | Value type for representing a single value in analysis data objects |
   AnalysisDataFrameHeader AnalysisDataFrameHeader | Value type for storing frame-level information for analysis data |
   AnalysisDataPointSetRef AnalysisDataPointSetRef | Value type wrapper for non-mutable access to a set of data column values |
   AnalysisDataFrameRef AnalysisDataFrameRef | Value type wrapper for non-mutable access to a data frame |
   IAnalysisDataModule IAnalysisDataModule | Interface for a module that gets notified whenever data is added |
   AnalysisDataModuleSerial AnalysisDataModuleSerial | Convenience base class for serial analysis data modules |
   AnalysisDataModuleParallel AnalysisDataModuleParallel | Convenience base class for parallel analysis data modules |
   AnalysisDataAverageModule AnalysisDataAverageModule | Data module for independently averaging each column in input data |
   AnalysisDataFrameAverageModule AnalysisDataFrameAverageModule | Data module for averaging of columns for each frame |
   AnalysisDataDisplacementModule AnalysisDataDisplacementModule | Data module for calculating displacements |
   AnalysisHistogramSettingsInitializer AnalysisHistogramSettingsInitializer | Provides "named parameter" idiom for constructing histograms |
   AnalysisHistogramSettings AnalysisHistogramSettings | Contains parameters that specify histogram bin locations |
   AbstractAverageHistogram AbstractAverageHistogram | Base class for representing histograms averaged over frames |
   AnalysisDataSimpleHistogramModule AnalysisDataSimpleHistogramModule | Data module for per-frame histograms |
   AnalysisDataWeightedHistogramModule AnalysisDataWeightedHistogramModule | Data module for per-frame weighted histograms |
   AnalysisDataBinAverageModule AnalysisDataBinAverageModule | Data module for bin averages |
   AnalysisDataLifetimeModule AnalysisDataLifetimeModule | Data module for computing lifetime histograms for columns in input data |
   AnalysisDataPlotSettings AnalysisDataPlotSettings | Common settings for data plots |
   AbstractPlotModule AbstractPlotModule | Abstract data module for writing data into a file |
   AnalysisDataPlotModule AnalysisDataPlotModule | Plotting module for straightforward plotting of data |
   AnalysisDataVectorPlotModule AnalysisDataVectorPlotModule | Plotting module specifically for data consisting of vectors |
   StaticLog2 StaticLog2 | Evaluate log2(n) for integer n statically at compile time |
   StaticLog2< 1 > StaticLog2< 1 > | Specialization of StaticLog2<n> for n==1 |
   StaticLog2< 0 > StaticLog2< 0 > | Specialization of StaticLog2<n> for n==0 |
   AbstractOption AbstractOption | Abstract base class for specifying option properties |
   OptionTemplate OptionTemplate | Templated base class for constructing concrete option settings classes |
   OptionInfo OptionInfo | Gives information and allows modifications to an option after creation |
   BooleanOption BooleanOption | Specifies an option that provides boolean values |
   IntegerOption IntegerOption | Specifies an option that provides integer values |
   Int64Option Int64Option | Specifies an option that provides 64-bit integer values |
   DoubleOption DoubleOption | Specifies an option that provides floating-point (double) values |
   FloatOption FloatOption | Specifies an option that provides floating-point (float) values |
   StringOption StringOption | Specifies an option that provides string values |
   EnumOption EnumOption | Specifies an option that accepts an EnumerationArray of string values and writes the selected index into an enum variable |
   LegacyEnumOption LegacyEnumOption | Specifies an option that accepts enumerated string values and writes the selected index into an enum variable |
   BooleanOptionInfo BooleanOptionInfo | Wrapper class for accessing boolean option information |
   IntegerOptionInfo IntegerOptionInfo | Wrapper class for accessing integer option information |
   Int64OptionInfo Int64OptionInfo | Wrapper class for accessing 64-bit integer option information |
   DoubleOptionInfo DoubleOptionInfo | Wrapper class for accessing floating-point option information |
   FloatOptionInfo FloatOptionInfo | Wrapper class for accessing floating-point option information |
   StringOptionInfo StringOptionInfo | Wrapper class for accessing string option information |
   EnumOptionInfo EnumOptionInfo | Wrapper class for accessing enum option information |
   FileNameOption FileNameOption | Specifies an option that provides file names |
   FileNameOptionInfo FileNameOptionInfo | Wrapper class for accessing file name option information |
   FileNameOptionManager FileNameOptionManager | Handles interaction of file name options with global options |
   IOptionsBehavior IOptionsBehavior | Interface to provide extension points for options parsing |
   IOptionsContainer IOptionsContainer | Interface for adding input options |
   IOptionsContainerWithSections IOptionsContainerWithSections | Interface for adding input options with sections |
   IOptionManager IOptionManager | Base class for option managers |
   Options Options | Collection of options |
   TimeUnitManager TimeUnitManager | Provides common functionality for time unit conversions |
   TimeUnitBehavior TimeUnitBehavior | Options behavior to add a time unit option |
   PotentialPointData PotentialPointData | Structure to hold the results of IRestraintPotential::evaluate() |
   IRestraintPotential IRestraintPotential | Interface for Restraint potentials |
   AnalysisNeighborhoodPositions AnalysisNeighborhoodPositions | Input positions for neighborhood searching |
   AnalysisNeighborhood AnalysisNeighborhood | Neighborhood searching for analysis tools |
   AnalysisNeighborhoodPair AnalysisNeighborhoodPair | Value type to represent a pair of positions found in neighborhood searching |
   AnalysisNeighborhoodSearch AnalysisNeighborhoodSearch | Initialized neighborhood search with a fixed set of reference positions |
   AnalysisNeighborhoodPairSearch AnalysisNeighborhoodPairSearch | Initialized neighborhood pair search with a fixed set of positions |
   Selection Selection | Provides access to a single selection |
   SelectionPosition SelectionPosition | Provides access to information about a single selected position |
   SelectionCollection SelectionCollection | Collection of selections |
   SelectionTopologyProperties SelectionTopologyProperties | Describes topology properties required for selection evaluation |
   SelectionOption SelectionOption | Specifies an option that provides selection(s) |
   SelectionOptionInfo SelectionOptionInfo | Wrapper class for accessing and modifying selection option information |
   ITopologyProvider ITopologyProvider | Provides topology information to SelectionOptionBehavior |
   SelectionOptionBehavior SelectionOptionBehavior | Options behavior to allow using SelectionOptions |
   SelectionOptionManager SelectionOptionManager | Handles interaction of selection options with other options and user input |
   RangePartitioning RangePartitioning | Division of a range of indices into consecutive blocks |
   TrajectoryAnalysisModuleData TrajectoryAnalysisModuleData | Base class for thread-local data storage during trajectory analysis |
   TrajectoryAnalysisModule TrajectoryAnalysisModule | Base class for trajectory analysis modules |
   TrajectoryAnalysisSettings TrajectoryAnalysisSettings | Trajectory analysis module configuration object |
   TrajectoryAnalysisCommandLineRunner TrajectoryAnalysisCommandLineRunner | Runner for command-line trajectory analysis tools |
   TopologyInformation TopologyInformation | Topology information available to a trajectory analysis module |
   EnumerationIterator EnumerationIterator | Allows iterating sequential enumerators |
   EnumerationWrapper EnumerationWrapper | Allows constructing iterators for looping over sequential enumerators |
   ExceptionInfo ExceptionInfo | Stores additional context information for exceptions |
   ExceptionInitializer ExceptionInitializer | Provides information for Gromacs exception constructors |
   GromacsException GromacsException | Base class for all exception objects in Gromacs |
   FileIOError FileIOError | Exception class for file I/O errors |
   UserInputError UserInputError | Exception class for user input errors |
   InvalidInputError InvalidInputError | Exception class for situations where user input cannot be parsed/understood |
   InconsistentInputError InconsistentInputError | Exception class for situations where user input is inconsistent |
   ToleranceError ToleranceError | Exception class when a specified tolerance cannot be achieved |
   SimulationInstabilityError SimulationInstabilityError | Exception class for simulation instabilities |
   InternalError InternalError | Exception class for internal errors |
   APIError APIError | Exception class for incorrect use of an API |
   RangeError RangeError | Exception class for out-of-range values or indices |
   NotImplementedError NotImplementedError | Exception class for use of an unimplemented feature |
   ParallelConsistencyError ParallelConsistencyError | Exception class for use when ensuring that MPI ranks to throw in a coordinated fashion |
   ModularSimulatorError ModularSimulatorError | Exception class for modular simulator |
   FlagsTemplate FlagsTemplate | Template class for typesafe handling of combination of flags |
   InstallationPrefixInfo InstallationPrefixInfo | Provides information about installation prefix (see IProgramContext::installationPrefix()) |
   IProgramContext IProgramContext | Provides context information about the program that is calling the library |
   StringFormatter StringFormatter | Function object that wraps a call to formatString() that expects a single conversion argument, for use with algorithms |
   IdentityFormatter IdentityFormatter | Function object to implement the same interface as StringFormatter to use with strings that should not be formatted further |
   EqualCaseInsensitive EqualCaseInsensitive | Function object for comparisons with equalCaseInsensitive |
   TextLineWrapperSettings TextLineWrapperSettings | Stores settings for line wrapping |
   TextLineWrapper TextLineWrapper | Wraps lines to a predefined length |
  Nbnxm Nbnxm | Namespace for non-bonded kernels |
   EnergyFunctionProperties EnergyFunctionProperties | Set of boolean constants mimicking preprocessor macros |
  AmdPackedFloat3 AmdPackedFloat3 | Special packed Float3 flavor to help compiler optimizations on AMD CDNA2 devices |
  AtomIterator AtomIterator | Object that allows looping over all atoms in an mtop |
  AtomProperties AtomProperties | Holds all the atom property information loaded |
  AtomProxy AtomProxy | Proxy object returned from AtomIterator |
  AtomRange AtomRange | Range over all atoms of topology |
  AtomsAdjacencyListElement AtomsAdjacencyListElement | Helper type for discovering coupled constraints |
  BalanceRegion BalanceRegion | Object that describes a DLB balancing region |
  BondedInteraction BondedInteraction | Information about single bonded interaction |
  BondedInteractionList BondedInteractionList | Accumulation of different bonded types for preprocessing |
  CheckpointHeaderContents CheckpointHeaderContents | Header explaining the context of a checkpoint file |
  DDBalanceRegionHandler DDBalanceRegionHandler | Manager for starting and stopping the dynamic load balancing region |
  DeviceInformation DeviceInformation | Platform-dependent device information |
  DeviceStream DeviceStream | Declaration of platform-agnostic device stream/queue |
  df_history_t df_history_t | Free-energy sampling history struct |
  ekinstate_t ekinstate_t | Struct used for checkpointing only |
  EwaldBoxZScaler EwaldBoxZScaler | Class to handle box scaling for Ewald and PME |
  FEPStateValue FEPStateValue | Template wrapper struct for particle FEP state values |
  ForceHelperBuffers ForceHelperBuffers | Helper force buffers for ForceOutputs |
  ForeignLambdaTerms ForeignLambdaTerms | Accumulates free-energy foreign lambda energies and dH/dlamba |
  gmx_ana_index_t gmx_ana_index_t | Stores a single index group |
  gmx_ana_indexmap_t gmx_ana_indexmap_t | Data structure for calculating index group mappings |
  gmx_ana_pos_t gmx_ana_pos_t | Stores a set of positions together with their origins |
  gmx_cache_protect_t gmx_cache_protect_t | Cache-line protection buffer |
  gmx_cmap_t gmx_cmap_t | CMAP grids |
  gmx_cmapdata_t gmx_cmapdata_t | CMAP data for a single grid |
  gmx_domdec_constraints_t gmx_domdec_constraints_t | Struct used during constraint setup with domain decomposition |
  gmx_enerdata_t gmx_enerdata_t | Struct for accumulating all potential energy terms and some kinetic energy terms |
  gmx_ffparams_t gmx_ffparams_t | Struct that holds all force field parameters for the simulated system |
  gmx_ga2la_t gmx_ga2la_t | Global to local atom mapping |
   Entry Entry | Structure for the local atom info |
  gmx_localtop_t gmx_localtop_t | The fully written out topology for a domain over its lifetime |
  gmx_molblock_t gmx_molblock_t | Block of molecules of the same type, used in gmx_mtop_t |
  gmx_moltype_t gmx_moltype_t | Molecules type data: atoms, interactions and exclusions |
  gmx_multisim_t gmx_multisim_t | Coordinate multi-simulation resources for mdrun |
  gmx_nodecomm_t gmx_nodecomm_t | Settings and communicators for two-step communication: intra + inter-node |
  gmx_sparsematrix gmx_sparsematrix | Sparse matrix storage format |
  GpuRegionTimerImpl GpuRegionTimerImpl | The OpenCL implementation of the GPU code region timing. With OpenCL, one has to use cl_event handle for each API call that has to be timed, and accumulate the timing afterwards. As we would like to avoid overhead on API calls, we only query and accumulate cl_event timing at the end of time steps, not after the API calls. Thus, this implementation does not reuse a single cl_event for multiple calls, but instead maintains an array of cl_events to be used within any single code region. The array size is fixed at a small but sufficiently large value for the number of cl_events that might contribute to a timer region, currently 10 |
  GpuRegionTimerWrapper GpuRegionTimerWrapper | This is a GPU region timing wrapper class. It allows for host-side tracking of the accumulated execution timespans in GPU code (measuring kernel or transfers duration). It also partially tracks the correctness of the timer state transitions, as far as current implementation allows (see TODO in getLastRangeTime() for a disabled check). Internally it uses GpuRegionTimerImpl for measuring regions |
  history_t history_t | History information for NMR distance and orientation restraints |
  IListIterator IListIterator | Object that allows looping over all atoms in an mtop |
  IListProxy IListProxy | Proxy object returned from IListIterator |
  IListRange IListRange | Range over all interaction lists of topology |
  IndexGroup IndexGroup | An index group consisting or a name and list of atom indices |
  InteractionDefinitions InteractionDefinitions | Struct with list of interaction parameters and lists of interactions |
  InteractionList InteractionList | List of listed interactions |
  InteractionListHandle InteractionListHandle | Type for returning a list of InteractionList references |
  InteractionOfType InteractionOfType | Describes an interaction of a given type, plus its parameters |
  InteractionsOfType InteractionsOfType | A set of interactions of a given type (found in the enumeration in ifunc.h), complete with atom indices and force field function parameters |
  JClusterList JClusterList | Simple j-cluster list |
  ListedForces ListedForces | Class for calculating listed interactions, uses OpenMP parallelization |
  LocalSettleData LocalSettleData | Local settle data |
  MoleculeBlockIndices MoleculeBlockIndices | Indices for a gmx_molblock_t, derived from other gmx_mtop_t contents |
  MoleculeInformation MoleculeInformation | Holds the molecule information during preprocessing |
  MoleculePatch MoleculePatch | Block to modify individual residues |
  MoleculePatchDatabase MoleculePatchDatabase | A set of modifications to apply to atoms |
  NbnxmKernel NbnxmKernel | Class name for NBNXM kernel |
  nbnxn_cj_packed_t nbnxn_cj_packed_t | Packed j-cluster list element |
  nbnxn_cj_t nbnxn_cj_t | This is the actual cluster-pair list j-entry |
  nbnxn_cycle_t nbnxn_cycle_t | Local cycle count struct for profiling |
  nbnxn_excl_t nbnxn_excl_t | Struct for storing the atom-pair interaction bits for a cluster pair in a GPU pairlist |
  nbnxn_im_ei_t nbnxn_im_ei_t | Interaction data for a j-group for one warp |
  nbnxn_sci_t nbnxn_sci_t | Grouped pair-list i-unit |
  NbnxnPairlistCpu NbnxnPairlistCpu | Cluster pairlist type for use on CPUs |
  NbnxnPairlistCpuWork NbnxnPairlistCpuWork | Working data for the actual i-supercell during pair search |
   IClusterData IClusterData | Struct for storing coordinates and bounding box for an i-entry during search |
  nonbonded_verlet_t nonbonded_verlet_t | Top-level non-bonded data structure for the Verlet-type cut-off scheme |
  NumPmeDomains NumPmeDomains | Struct for passing around the number of PME domains |
  ObservablesHistory ObservablesHistory | Observables history, for writing/reading to/from checkpoint file |
  PackedJClusterList PackedJClusterList | Packed j-cluster list |
  PairlistSets PairlistSets | Contains sets of pairlists |
  PairSearch PairSearch | Main pair-search struct, contains the grid(s), not the pair-list(s) |
  PairsearchWork PairsearchWork | Thread-local work struct, contains working data for Grid |
  PartialDeserializedTprFile PartialDeserializedTprFile | Contains the partly deserialized contents of a TPR file |
  PbcAiuc PbcAiuc | Compact and ordered version of the PBC matrix |
  PdbAtomEntry PdbAtomEntry | Contains information for a single particle in a PDB file |
  PmeGpuProgram PmeGpuProgram | Stores PME data derived from the GPU context or devices |
  PmeSolve PmeSolve | Class for solving PME for Coulomb and LJ |
  PpRanks PpRanks | Contains information about the PP ranks that partner this PME rank |
  PreprocessingAtomTypes PreprocessingAtomTypes | Storage of all atom types used during preprocessing of a simulation input |
  PreprocessingBondAtomType PreprocessingBondAtomType | Storage for all bonded atomtypes during simulation preprocessing |
  PreprocessResidue PreprocessResidue | Information about preprocessing residues |
  ReplicaExchangeParameters ReplicaExchangeParameters | The parameters for the replica exchange algorithm |
  SettleWaterTopology SettleWaterTopology | Composite data for settle initialization |
  SimulationGroups SimulationGroups | Contains the simulation atom groups |
  SimulationInputHandle SimulationInputHandle | Owning handle to a SimulationInput object |
  SimulationParticle SimulationParticle | Single particle in a simulation |
  StringTable StringTable | A class to store strings for lookup |
  StringTableBuilder StringTableBuilder | Builds a memory efficient storage for strings of characters |
  StringTableEntry StringTableEntry | Helper class to access members in StringTable |
  SystemMomenta SystemMomenta | System momenta used with with the box deform option |
  SystemMomentum SystemMomentum | The momentum and mass of the whole system |
  t_filenm t_filenm | File name option definition for C code |
  t_idef t_idef | Deprecated interation definitions, used in t_topology |
  t_ilist t_ilist | Deprecated list of listed interactions |
  t_interaction_function t_interaction_function | Interaction function |
  t_iparams t_iparams | Union of various interaction paramters |
  t_mapping t_mapping | Maps an XPM element to an RGB color and a string description |
  t_matrix t_matrix | A matrix of integers, plus supporting values, such as used in XPM output |
  t_mdatoms t_mdatoms | Declares mdatom data structure |
  t_oriresdata t_oriresdata | Orientation restraining stuff |
  t_pargs t_pargs | Command-line argument definition for C code |
  t_pbc t_pbc | Structure containing info on periodic boundary conditions |
  t_state t_state | The microstate of the system |
  t_symbuf t_symbuf | Legacy symbol table entry as linked list |
  t_xpmelmt t_xpmelmt | Models an XPM element |
  TpxFileHeader TpxFileHeader | First part of the TPR file structure containing information about the general aspect of the system |
  wallcc_t wallcc_t | Counters for an individual wallcycle timing region |
  WarningHandler WarningHandler | General warning handling object |
  WaterMolecule WaterMolecule | Indices of atoms in a water molecule |
 1.8.5
1.8.5